DREAM 2021: Disentangled Representations for Efficient Algorithms for Medical data
A MICCAI 2021 (Virtual) Tutorial by Sotirios A. Tsaftaris, Alison Q. O’Neil, Spyridon Thermos, Xiao Liu and Pedro Sanchez.

Date
MICCAI day 1 - September 27, 2021 - 14h-18h [UTC time]
Accompanying Material
 |  |  |  |
- The contents of this tutorial have been compiled into a paper published at the Medical Image Analysis journal;
- A github repository summarizes the codebases of several important works in the area;
- The videos of the presentations are available on our YouTube channel, find the playlist here;
- The tutorial’s slide deck in pdf is also available here.
@article{liu2022disentangled,
title = {Learning disentangled representations in the imaging domain},
journal = {Medical Image Analysis},
volume = {80},
pages = {102516},
year = {2022},
issn = {1361-8415},
doi = {https://doi.org/10.1016/j.media.2022.102516},
author = {Xiao Liu and Pedro Sanchez and Spyridon Thermos and Alison Q. O’Neil and Sotirios A. Tsaftaris},
}
Outline
The deep learning (DL) paradigm has been widely adopted in almost all domains of image analysis as an alternative to traditional handcrafted techniques. However, the majority of deep neural networks rely on the existence of significant amounts of training data that are not always readily available. Medical image analysis is a characteristic example of a field where the difficulty and expense of acquiring and annotating data prohibit the true exploitation of the deep learning potential.
Disentangled representation learning has been proposed as an approach to encode generic and explainable data representations through separating out underlying explanatory factors. Interestingly, this can be achieved with limited or no annotations. A general and explainable representation can be readily fine-tuned for new target tasks using modest amounts of data. This alleviation of the data and annotation requirements offers tantalising prospects for tractable and affordable healthcare, while the explainability of disentangled representations increase their suitability for real-world human-controlled applications.
In this half-day tutorial, a satellite event in conjunction with MICCAI 2021 (Virtual), we present an overview of representation learning, focusing on disentangled representation learning and criteria, as well as on the connection between disentangled representations and causal mechanisms. Finally, we discuss about possible applications in the medical imaging field and existing open-ended challenges. This tutorial is a follow-up of the DREAM @ MICCAI 2020 which was extremely well received and attended by around 200 members of the MICCAI community.
Program


Learning Objectives
- Understand representation space and why (in)variance matters
- Understand the theories of information bottleneck and compositionality
- Learn the different objectives in achieving disentanglement
- Appreciate the inductive biases introduced by network design choices
- Appreciate when disentanglement is useful in practice
Motivation
Imagine that we want to develop a system that localises the heart in MRI and CT images. This system will need to be robust to any changes in imaging, the scanner, noise, and critically anatomical and pathological variation. The current paradigm with deep learning is that we must present to the system as many examples as possible to make it robust and learn what is nuisance (e.g. noise and imaging differences) as opposed to what matters (e.g. the location of the heart). [A similar argument can be made for organs in a CAI setting.]
Clearly this is not sustainable and leads to poor performance. Disentangled learning can help because it allows us to learn latent factors that can describe what we see in the data. In the example on the right, we can imagine a latent factor that one learns a change in pose (or in the patient for the cardiac example), and another factor for the change in a car’s colour (or a different scanner). Surprisingly we do not always need annotated data to achieve this. Moreover, it has been shown that disentangled representations are privacy preserving; can offer explainability and interpretability; and can generalise to new tasks (meta-learning) and to new data sources with less effort.
We should (re)appreciate that machine learning (ML) is not simply a functional mapping between input and output, but one that maps input data to manifolds and then decisions/tasks. This return to principled design will allow us to evolve and propose efficient solutions that are robust to new applications and shorten the path to clinical translation.
Learning suitable representations is key in ML. In fact, the ML community has the dedicated International Conference on Learning Representations (ICLR). Disentangled learning is considered a hot area: a dedicated challenge run at NeurIPS 2019. According to Google Scholar trends, the number of papers in major ML conferences doubles yearly . Scientific American, a prestigious periodical in popular science, had a description of disentangled representation learning in May 2019 (Machine Learning Gets a Bit More Humanlike). Disentangled learning appeared in MICCAI 5 years ago in a couple of papers, but now has tripled in appearance. Thus, it is timely to formally introduce this important area of research in our community. A google scholar search outputs the following number of disentanglement publications per year:
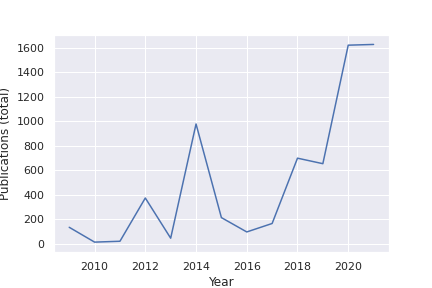
Query: “learning AND (disentangled OR disentanglement) source:Arxiv”
Applications
But is disentanglement relevant to real-life applications? We will answer this question reporting details of exemplar methods that exploit disentanglement to improve challenging tasks in computer vision and medical image analysis.
Teachers
Prof. Sotirios A Tsaftaris (Sotos) is Chair in Machine Learning and Computer Vision with the University of Edinburgh and is also the Canon Medical/Royal Academy of Engineering Research Chair in Healthcare AI. Sotos leads a group where several young researchers work in machine learning and computer vision. He obtained his PhD in 2006 from Northwestern University USA and has held several academic positions in USA, Italy and UK. Sotos’s research expertise is in representation learning.
Dr Alison Q O’Neil (Alison) is a Senior Scientist in the AI Research Team at Canon Medical Research Europe and Honorary Research Fellow at the University of Edinburgh. She obtained her EngD at Canon Medical Research Europe in affiliation with Heriot-Watt University, and now leads a team of scientists and research students working on machine learning techniques for industrial healthcare applications – including applications in medical imaging, natural language processing, and electronic health record (EHR) data. Alison will bring an industry perspective to the tutorial and also talk about disentanglement in other data forms (e.g. text).
Dr. Spyridon Thermos (Spiros) is a postdoc at UoE. Spiros expertise lies in disentangled representation learning, disentanglement evaluation and conditional image synthesis. He will present metrics on disentanglement and how to measure the entanglement between tensors and latent representations.
Mr. Xiao Liu is a 2nd year PhD student at UoE. His research interests include cardiac image segmentation, disentangled representation learning and domain generalization. He will present the role of decoders in learning representations and how disentanglement can help with domain adaptation and generalisation.
Mr. Pedro Sanchez is a 1st year PhD student working on disentanglement and causal learning. Previously, he worked at Canon Medical with medical image analysis and machine learning. He completed his MSc in biomedical engineering at the University of Strasbourg. He will present the role of disentanglement in learning causal mechanisms.
Support
Generously supported by Canon Medical Research Europe, the Royal Academy of Engineering and the School of Engineering.

